Mozilla wants to make it easier to create, view, and replicate data visualizations on the web, and toward that end, it today unveiled Iodide, an “experimental tool” meant to help scientists and engineers write and share interactive documents using an iterative workflow. It’s currently in alpha, and available from GitHub in open source.
“In the last ten years, there has been an explosion of interest in ‘scientific computing’ and ‘data science’: that is, the application of computation to answer questions and analyze data in the natural and social sciences,” Brendan Colloran, staff data scientist at Mozilla, wrote in a blog post. “To address these needs, we’ve seen a renaissance in programming languages, tools, and techniques that help scientists and researchers explore and understand data and scientific concepts, and to communicate their findings. But to date, very few tools have focused on helping scientists gain unfiltered access to the full communication potential of modern web browsers.”
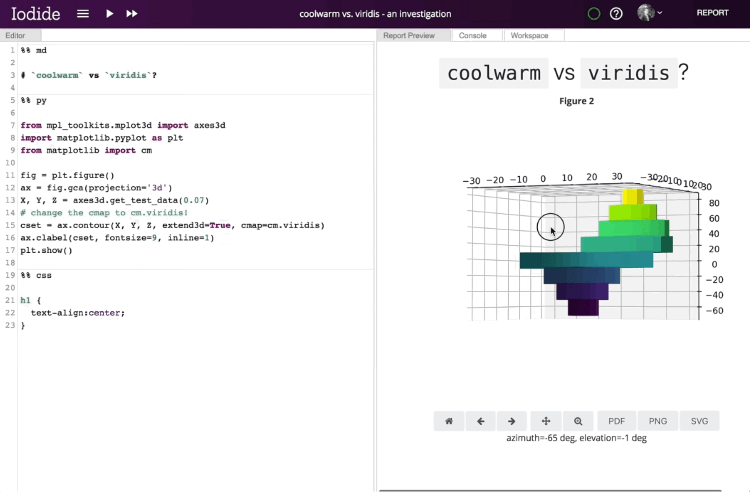
Within Iodide, users fill out pages with their content and use tools to modify the contributions they’ve made. The Explore view — the default view — consists of an editor for writing code, a console for viewing the output of that code, a workspace viewer for examining created variables, and a report preview pane. The editor, as with Jupyter and other popular open source report editors, allows authors to break code into segments (denoted with “%%” followed by a string indicating the programming language) that can be run independently of one another.
Iodide’s markdown format — JavaSscript MarkDown, or JSMD — currently supports code cells containing Javascript, CSS, Markdown (and HTML), Python, a “fetch” cell that streamlines resource loading, and a plugin chunk that adds new cell types. More are on the way, according to Colloran.
June 5th: The AI Audit in NYC
Join us next week in NYC to engage with top executive leaders, delving into strategies for auditing AI models to ensure fairness, optimal performance, and ethical compliance across diverse organizations. Secure your attendance for this exclusive invite-only event.
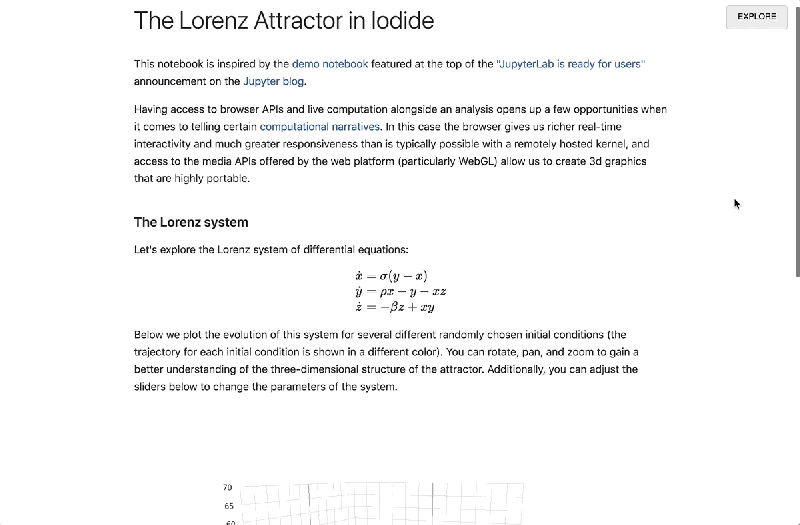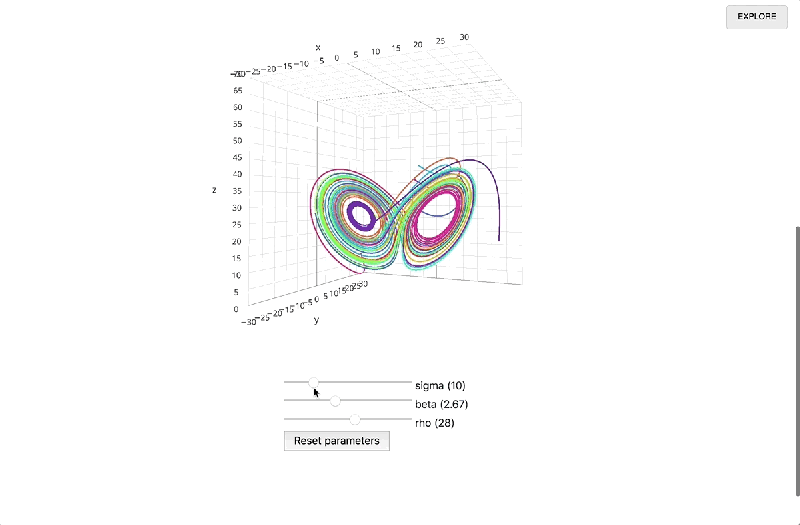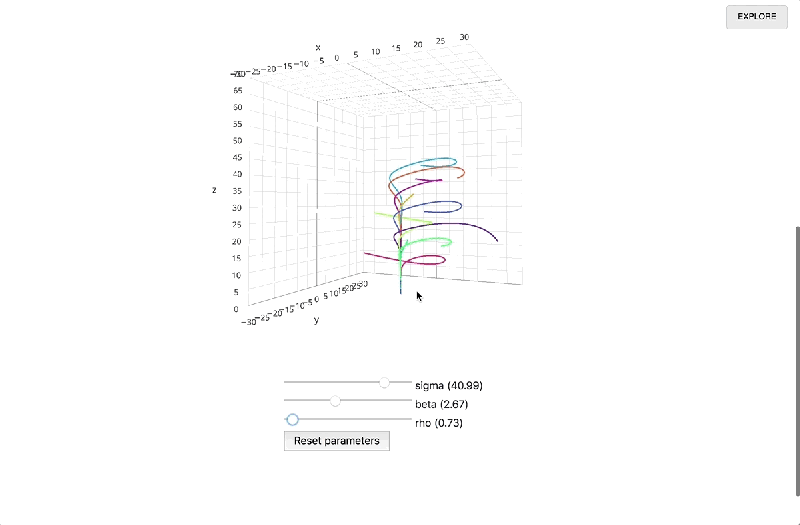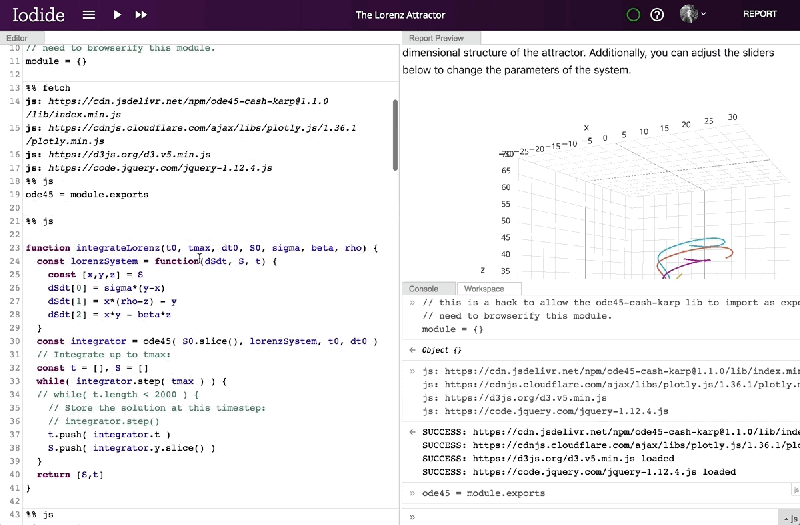
Once a report’s been finalized and published, a link to the interactive page can be shared with anyone on the web. Libraries are loaded via HTTP request, and computation happens in the browser alongside the presentation, obviating the need to call a language backend in another process and opening the possibility of 3D visualizations. Documents update in real time, and recipients who want to review the underlying code or fork it can drop into the Explore view with a single click.
Moreover, Iodide packs a Python science stack — Pyiodide — compiled to run on WebAssembly, a binary instruction format that allows web browsers to run code written in languages other than Javascript. Currently, it loads Numpy, Pandas, Matplotlib, Scipy, scikit-learn, and other libraries with only a slight performance penalty — Python runs about one to 12 times slower than native on Firefox and one to 16 times slower on Chrome, Mozilla says.
“Building Iodide in the web simplifies a number of the elements of workflow friction that we’ve encountered in other tools,” Colloran explains. “Sharing is simplified because the write-up and the code are available at the same URL rather than, say, pasting a link to a script in the footnotes of a Google Doc … And because browsers provide a compatibility layer, you don’t have to worry about notebook behavior being reproducible across computers and OSes.”
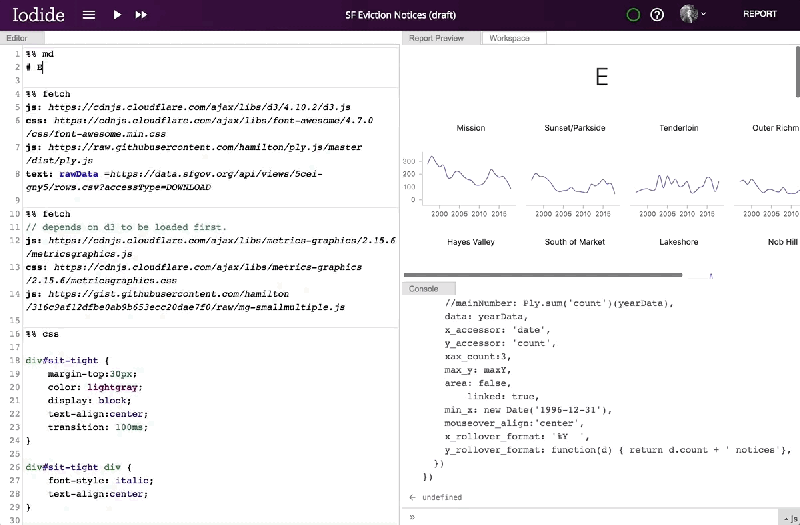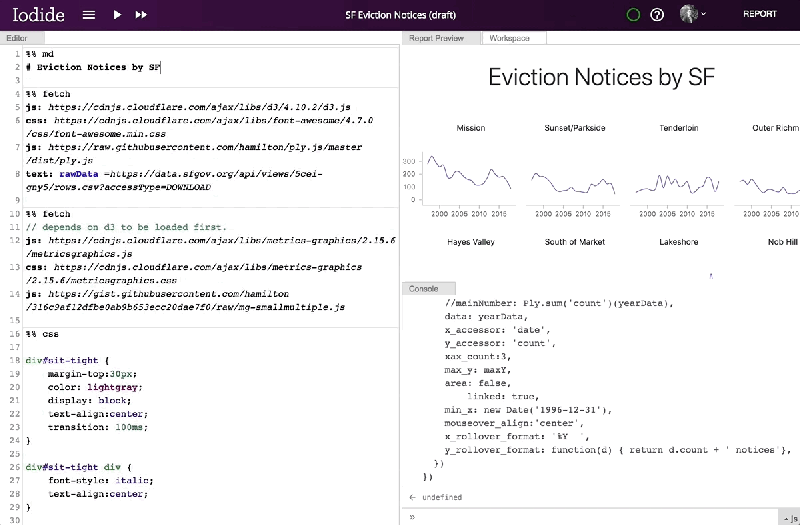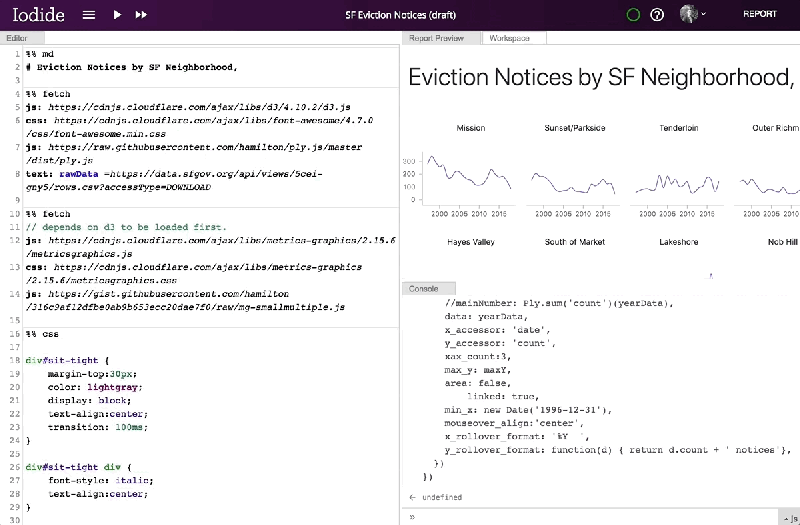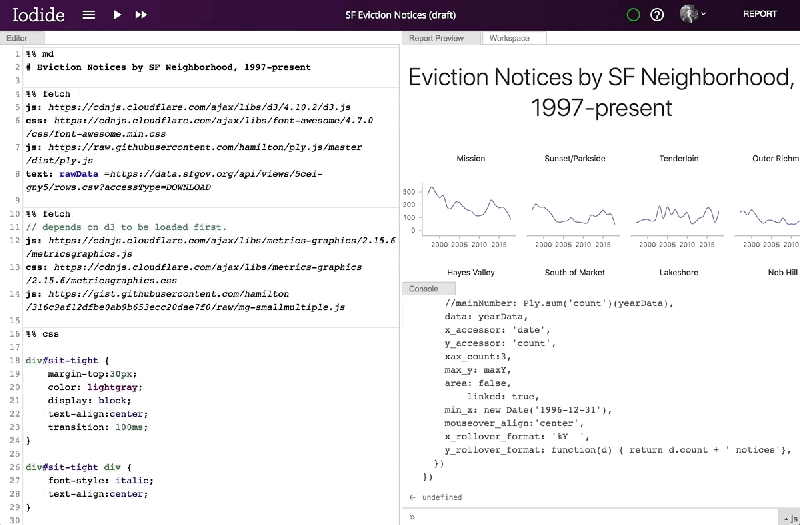
Mozilla has provided a server for saving and sharing Iodide notebooks, but notes that the notebooks themselves can be migrated to another server or exported as a bundle for sharing on other services like GitHub Pages or Netlify. And it says that, in the future, it hopes to add comment threads, the ability to suggest changes to users’ notebooks, simultaneous notebook editing, more languages, and a browser extension and APIs that would allow Iodide to work with client-side text editors.
“Iodide touches a wide variety of software disciplines, from modern frontend development to scientific computing to compilation and transpilation, so there are a lot of interesting things to do,” Colloran wrote. “We’re hoping that, despite the rough edges, if you squint at this you’ll be able to see the value of the concept, and that the feedback you give us will help us figure out where to go next.”