Watch all the Transform 2020 sessions on-demand here.
The recipe for proteins — large molecules consisting of amino acids that are the fundamental building block of tissues, muscles, hair, enzymes, antibodies, and other essential parts of living organisms — are encoded in DNA. It’s these genetic definitions that circumscribe their three-dimensional structure, which in turn determines their capabilities. Antibody proteins are shaped like a “Y,” for example, enabling them to latch onto viruses and bacteria, and collagen proteins are shaped like cords, which transmit tension between cartilage, bones, skin, and ligaments.
But protein “folding,” as it’s called, is notoriously difficult to suss out from a corresponding genetic sequence alone — DNA contains only information about chains of amino acid residues, and not those chains’ final form. In fact, scientists estimate that because of the incalculable number of interactions between the amino acids, it would take longer than 13.8 billion years to figure out all the possible configurations of a typical protein before identifying the right structure.
Fortunately, it’s a job fit for artificial intelligence (AI). Google subsidiary DeepMind this week announced AlphaFold, an AI system designed to predict protein structures more precisely than prior state-of-the-art solutions. It’s the product of two years of work, DeepMind researchers wrote in a blog post, and builds on years of genomics research.
“Over the past five decades, scientists have been able to determine shapes of proteins in labs using experimental techniques like cryo-electron microscopy, nuclear magnetic resonance, or X-ray crystallography, but each method depends on a lot of trial and error, which can take years and cost tens of thousands of dollars per structure,” the team wrote. “Fortunately, the field of genomics is quite rich in data, thanks to the rapid reduction in the cost of genetic sequencing. As a result, deep learning approaches to the prediction problem that rely on genomic data have become increasingly popular in the last few years.”
June 5th: The AI Audit in NYC
Join us next week in NYC to engage with top executive leaders, delving into strategies for auditing AI models to ensure fairness, optimal performance, and ethical compliance across diverse organizations. Secure your attendance for this exclusive invite-only event.
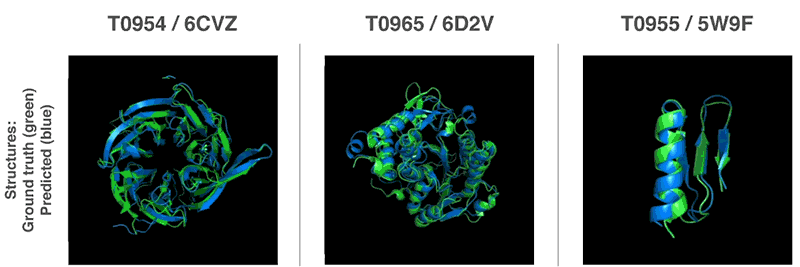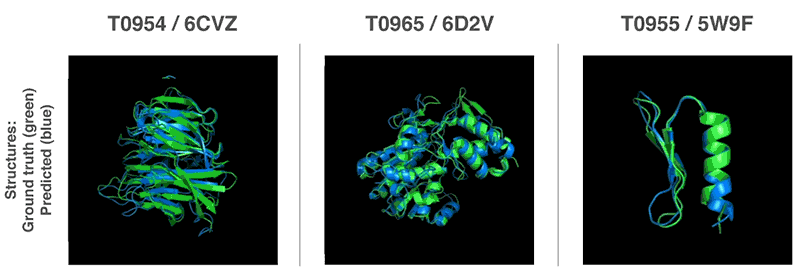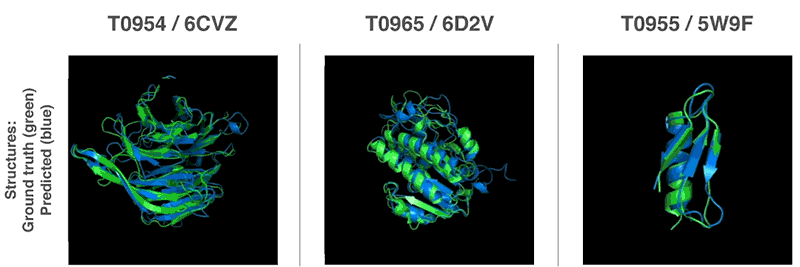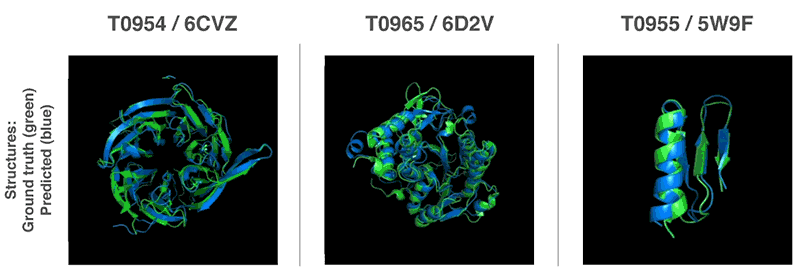
Above: Predicted protein structures compared to the ground truth.
The DeepMind team focused on the problem of modeling target shapes from scratch and used two methods to construct predictions of full protein structures. Specifically, their AI system’s deep neural networks — layers of mathematical functions that imitate the behavior of neurons in the human brain — are able to estimate the distance between pairs of amino acids and the angles between the chemical bonds that connect those amino acids.
One neural network in AlphaFold predicts the distribution of distances between pairs of amino acid residues in a protein by repeatedly placing pieces of a protein structure with new protein fragments. A generative model creates new fragments, which are used to continually improve precision. The probabilities are combined into a score that estimates the accuracy of a proposed structure, which a separate neural network evaluates using all distances in aggregate.
AlphaFold handily outguns competing solutions in terms of performance. It beat 98 competitors in the Critical Assessment of Structure Prediction (CASP) protein-folding competition in Cancun, where it successfuly predicted the structure of 25 out of 43 proteins. (The second-place system could only predict three protein structures.) More impressively, it managed to forecast its first protein structures in a matter of hours — magnitudes faster than previous systems.
Improving the scientific community’s understanding of protein folding could lead to more effective diagnoses and treatment of diseases such as Parkinson’s and Alzheimer’s, the DeepMind team noted, as these are believed to be caused by misfolded proteins. And it could aid in protein design, leading to protein-secreting bacteria that make wastewater biodegradable, for instance, and enzymes that can help manage pollutants such as plastic and oil.

“It’s exciting to see these early signs of progress in protein folding, demonstrating the utility of AI for scientific discovery,” the researchers wrote. “Even though there’s a lot more work to do before we’re able to have a quantifiable impact on treating diseases, managing the environment, and more, we know the potential is enormous. With a dedicated team focused on delving into how machine learning can advance the world of science, we’re looking forward to seeing the many ways our technology can make a difference.”


